Datasets
Contents
3.4. Datasets#
3.4.1. MIT-BIH Arrhythmia Database#
We use the MIT-BIH Arrhythmia Database [25] from PhysioNet [16]. The database contains 48 half-hour excerpts of two-channel ambulatory ECG recordings from 47 subjects. The recordings were digitized at 360 samples per second for both channels with 11-bit resolution over a 10mV range. The samples can be read in both digital (integer) form or as physical values (floating point) via the software provided by PhysioNet. We use the integer values in our experiments since our encoder is designed for integer arithmetic. We use the MLII signal (first channel) from each recording in our experiments.
3.4.1.1. Beats#
Each beat in the dataset is carefully annotated by a mutual agreement between two cardiologists. There were only a few beats that were very difficult to interpret and were left unclassified by the expert annotators. Different beat annotations found in the dataset and the number of their occurrences across all the 48 records are shown in Table 3.2.
sym |
meaning |
counts |
|
|---|---|---|---|
0 |
N |
normal beat |
75052 |
1 |
L |
left bundle branch block |
8075 |
2 |
R |
right bundle branch block |
7259 |
3 |
V |
premature ventricular contraction |
7130 |
4 |
/ |
paced beat |
7028 |
5 |
A |
atrial premature beat |
2546 |
6 |
f |
fusion of paced and normal beat |
982 |
7 |
F |
fusion of ventricular and normal beat |
803 |
8 |
j |
nodal (junctional) escape beat |
229 |
9 |
a |
aberrated atrial premature beat |
150 |
10 |
E |
ventricular escape beat |
106 |
11 |
J |
nodal (junctional) premature beat |
83 |
12 |
Q |
unclassifiable beat |
33 |
13 |
e |
atrial escape beat |
16 |
14 |
S |
supraventricular premature or ectopic beat (atrial or nodal) |
2 |
3.4.2. Sparsity#
In this section, we study the representation of ECG signals in different wavelet bases. The primary goal here is to understand the sparsity pattern of their representations in different bases. If a signal is suitably sparse on an appropriate basis (orthonormal or not), then such a basis can be used effectively for applying an appropriate signal compression method. Towards this, we study the sparsity properties of the wavelet representations of ECG signals in 106 different wavelets from 7 different families. The list of wavelets is provided in Table 3.3.
Family |
Wavelets |
|---|---|
haar |
haar |
db |
db1, db2, db3, db4, db5, db6, db7, db8, db9, db10, db11, db12, db13, db14, db15, db16, db17, db18, db19, db20, db21, db22, db23, db24, db25, db26, db27, db28, db29, db30, db31, db32, db33, db34, db35, db36, db37, db38 |
sym |
sym2, sym3, sym4, sym5, sym6, sym7, sym8, sym9, sym10, sym11, sym12, sym13, sym14, sym15, sym16, sym17, sym18, sym19, sym20 |
coif |
coif1, coif2, coif3, coif4, coif5, coif6, coif7, coif8, coif9, coif10, coif11, coif12, coif13, coif14, coif15, coif16, coif17 |
bior |
bior1.1, bior1.3, bior1.5, bior2.2, bior2.4, bior2.6, bior2.8, bior3.1, bior3.3, bior3.5, bior3.7, bior3.9, bior4.4, bior5.5, bior6.8 |
rbio |
rbio1.1, rbio1.3, rbio1.5, rbio2.2, rbio2.4, rbio2.6, rbio2.8, rbio3.1, rbio3.3, rbio3.5, rbio3.7, rbio3.9, rbio4.4, rbio5.5, rbio6.8 |
dmey |
dmey |
3.4.3. Wavelet Comparison Procedure#
The following procedure was employed to estimate the sparsity.
For each wavelet and each record
Split the record into blocks of
For each
Compute the
Sort the wavelet coefficients by magnitude in descending order.
Count the number of largest magnitude coefficients that capture 90% energy of the signal as the number
Put together the sparsity of each block into an array
Find the maximum, minimum, and mean values of the
Store the
A sample of
record |
rbio3.3 |
coif7 |
db14 |
coif16 |
haar |
coif2 |
db1 |
dmey |
|---|---|---|---|---|---|---|---|---|
222 |
395 |
226 |
217 |
226 |
311 |
212 |
311 |
219 |
231 |
121 |
96 |
104 |
133 |
128 |
80 |
128 |
100 |
207 |
368 |
210 |
211 |
218 |
341 |
234 |
341 |
218 |
208 |
288 |
165 |
178 |
193 |
248 |
174 |
248 |
172 |
111 |
283 |
164 |
175 |
189 |
245 |
164 |
245 |
169 |
217 |
128 |
95 |
107 |
132 |
131 |
80 |
131 |
96 |
223 |
108 |
84 |
91 |
133 |
111 |
70 |
111 |
88 |
219 |
98 |
86 |
97 |
135 |
90 |
66 |
90 |
87 |
3.4.4. The Biorthogonal 3.1 Wavelet#
Analyzing the data across all records and wavelets, we found that the “bior3.1” wavelet gives the best compressibility performance on average. A sample of the spread of k values from some records with bior3.1 wavelet can be seen in Table 3.5. Among this sample of records, one can see that record 117 appears to be one of the simplest ones as the maximum sparsity level is just 35 while record 200 appears to be one of the complex ones where the sparsity level goes as high as 137. We can also see that in all signals, there are segments that require very few coefficients (20 or less) for almost lossless representation. This is indicative of the fact that when the heart is behaving relatively stably, the ECG pattern is also less complex.
record |
||||
|---|---|---|---|---|
0 |
103 |
36.72 |
18 |
53 |
1 |
106 |
34.23 |
17 |
68 |
2 |
111 |
37.80 |
9 |
98 |
3 |
117 |
22.02 |
11 |
35 |
4 |
121 |
18.78 |
7 |
44 |
5 |
200 |
39.38 |
14 |
137 |
6 |
208 |
36.99 |
9 |
113 |
7 |
213 |
41.99 |
21 |
62 |
8 |
215 |
52.56 |
20 |
114 |
9 |
230 |
37.53 |
18 |
63 |
3.4.5. A Normal Sinus Rhythm#
A 1024 sample segment of a normal sinus rhythm in record 102 is depicted in Fig. 3.1. The wavelet coefficients are seen in Fig. 3.2. One can see that about 50 coefficients are needed to capture 99% of the signal energy. Another segment from record 100 in Fig. 3.3 with wavelet coefficients in Fig. 3.4 shows that the sparsity level for normal sinus rhythm can be as low as 21. We haven’t yet established the range of variation in the sparsity level for different types of sinus rhythms under the bior3.1 wavelet.
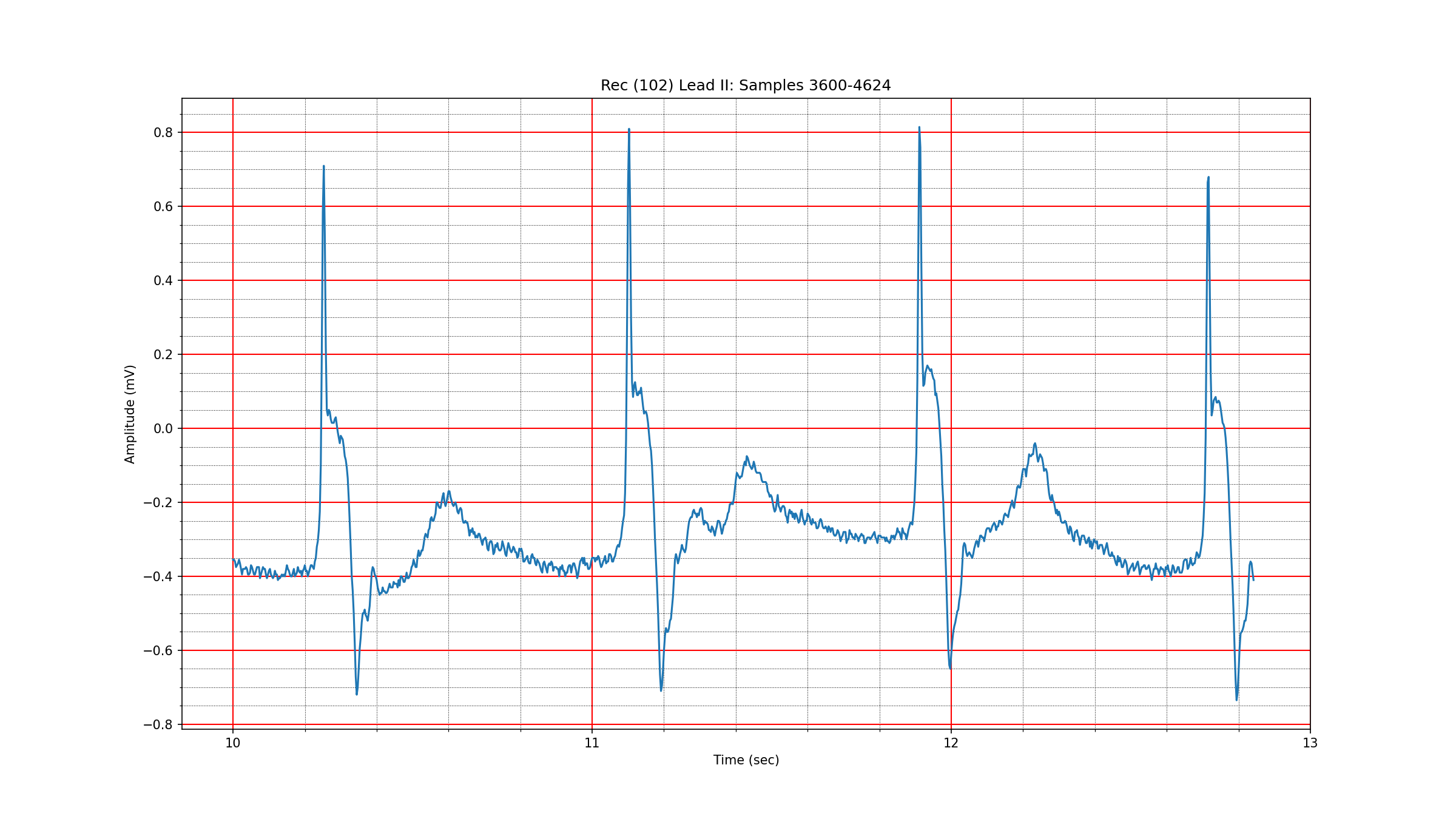
Fig. 3.1 A normal sinus rhythm in record 102#
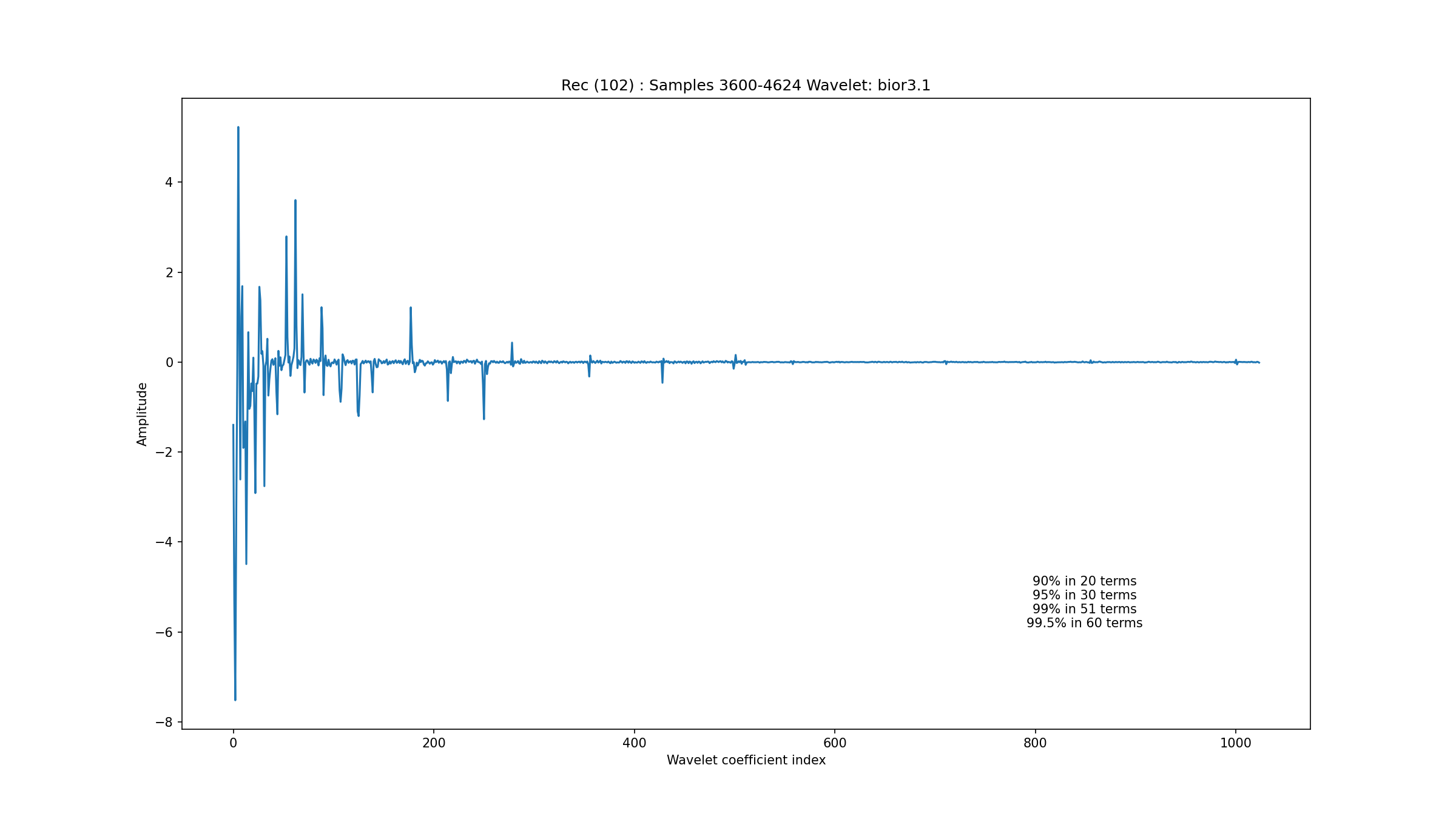
Fig. 3.2 Wavelet coefficients for the normal rhythm in record 102#
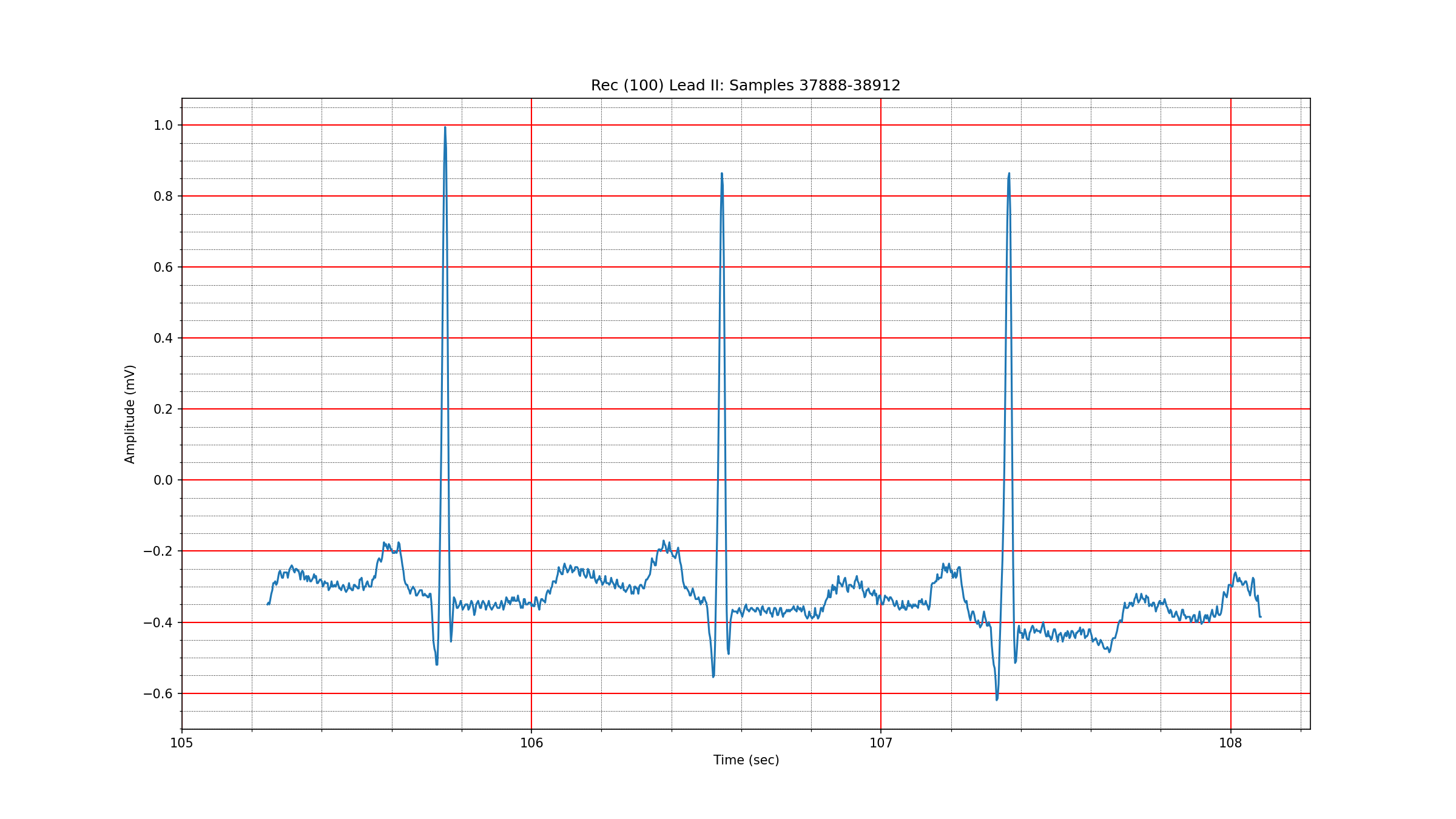
Fig. 3.3 A normal sinus rhythm in record 100#
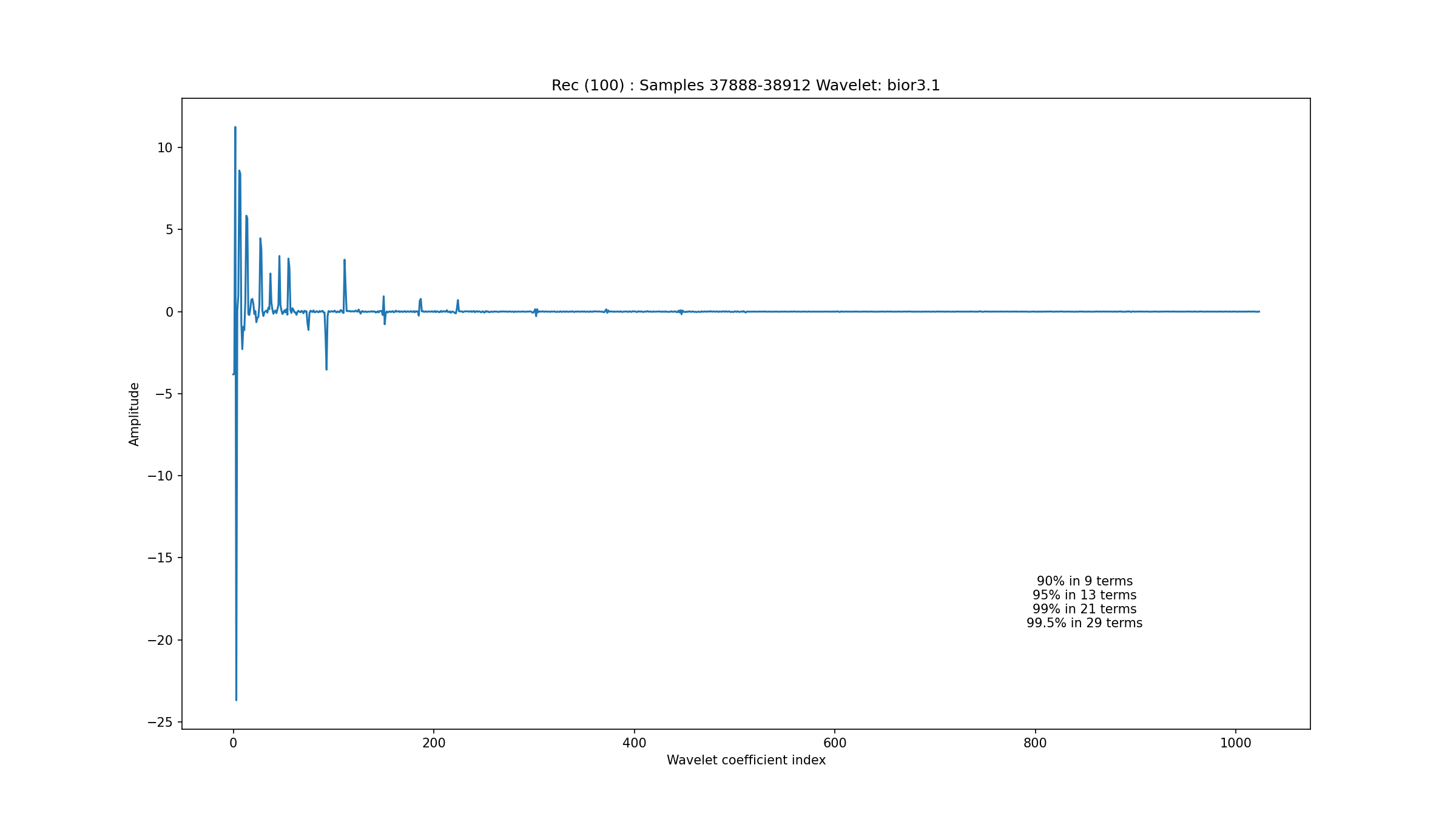
Fig. 3.4 Wavelet coefficients for the normal rhythm in record 100#
3.4.6. A Baseline Wander Segment#
Fig. 3.5 shows an excerpt of a baseline wander from record 121. It turns out that the wavelet representation for this signal is extremely sparse as seen in Fig. 3.6 with just 7 terms capturing 99% of the energy. One can see that most of the energy in this excerpt is coming from a DC component and a linear trend. This is why this signal has such a highly sparse representation. We noticed that very low sparsity levels tended to correspond to the presence of linear trends in the data.
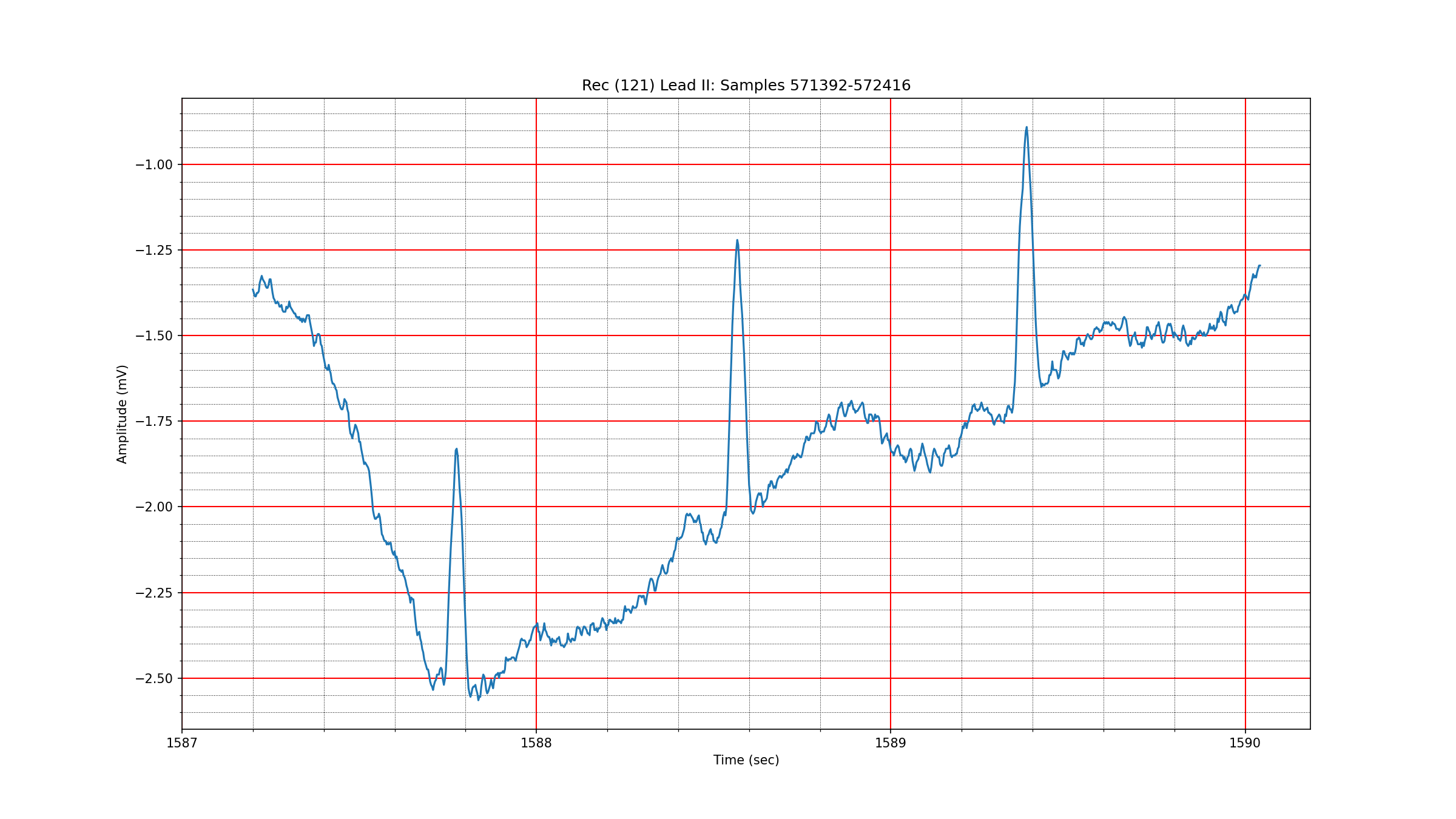
Fig. 3.5 A baseline wander in record 121#
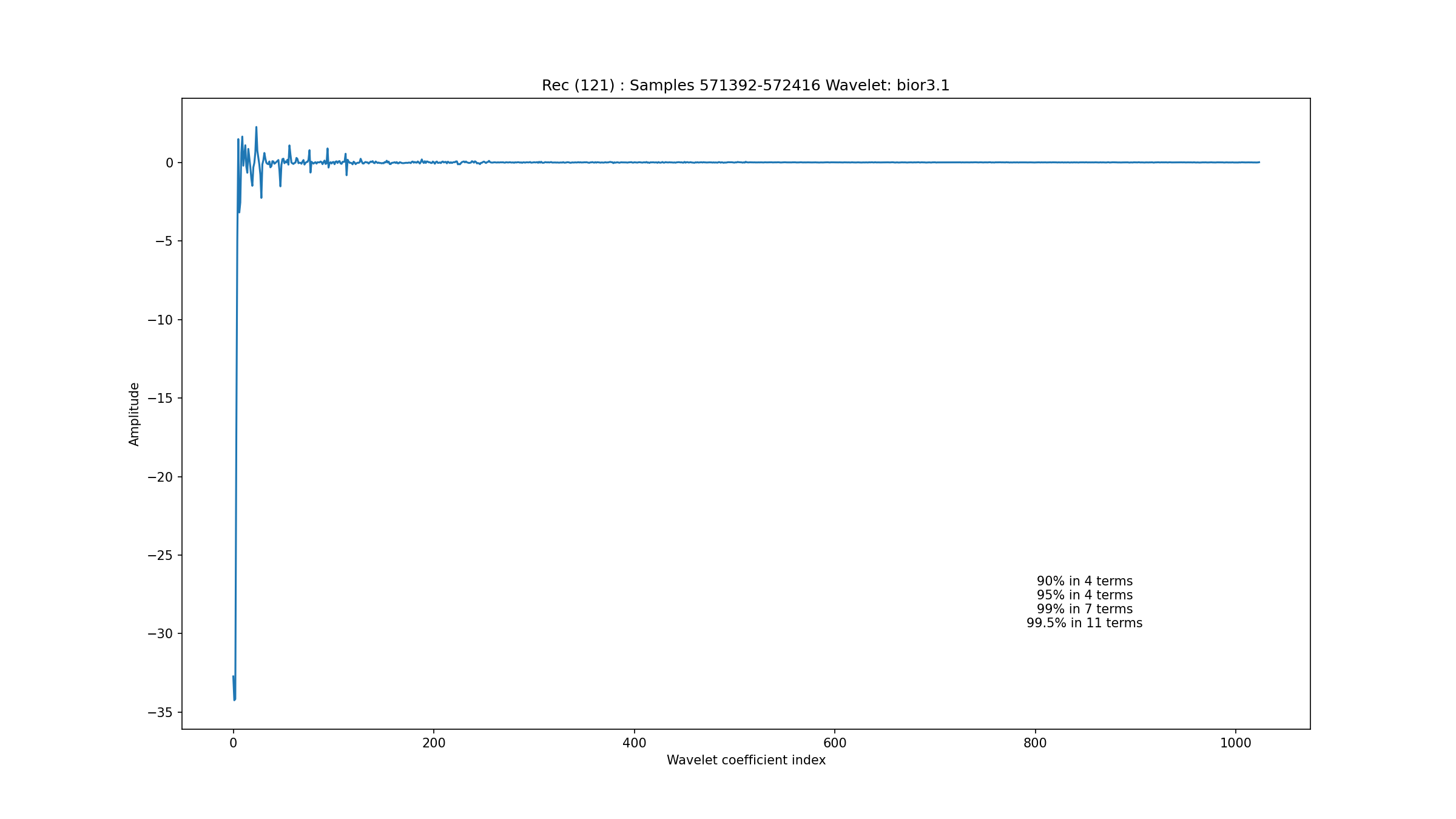
Fig. 3.6 Wavelet coefficients for baseline wander in record 121#
3.4.7. A Complex ECG Segment#
One of the most complex segments of ECG waveforms is found in the record 104
at around
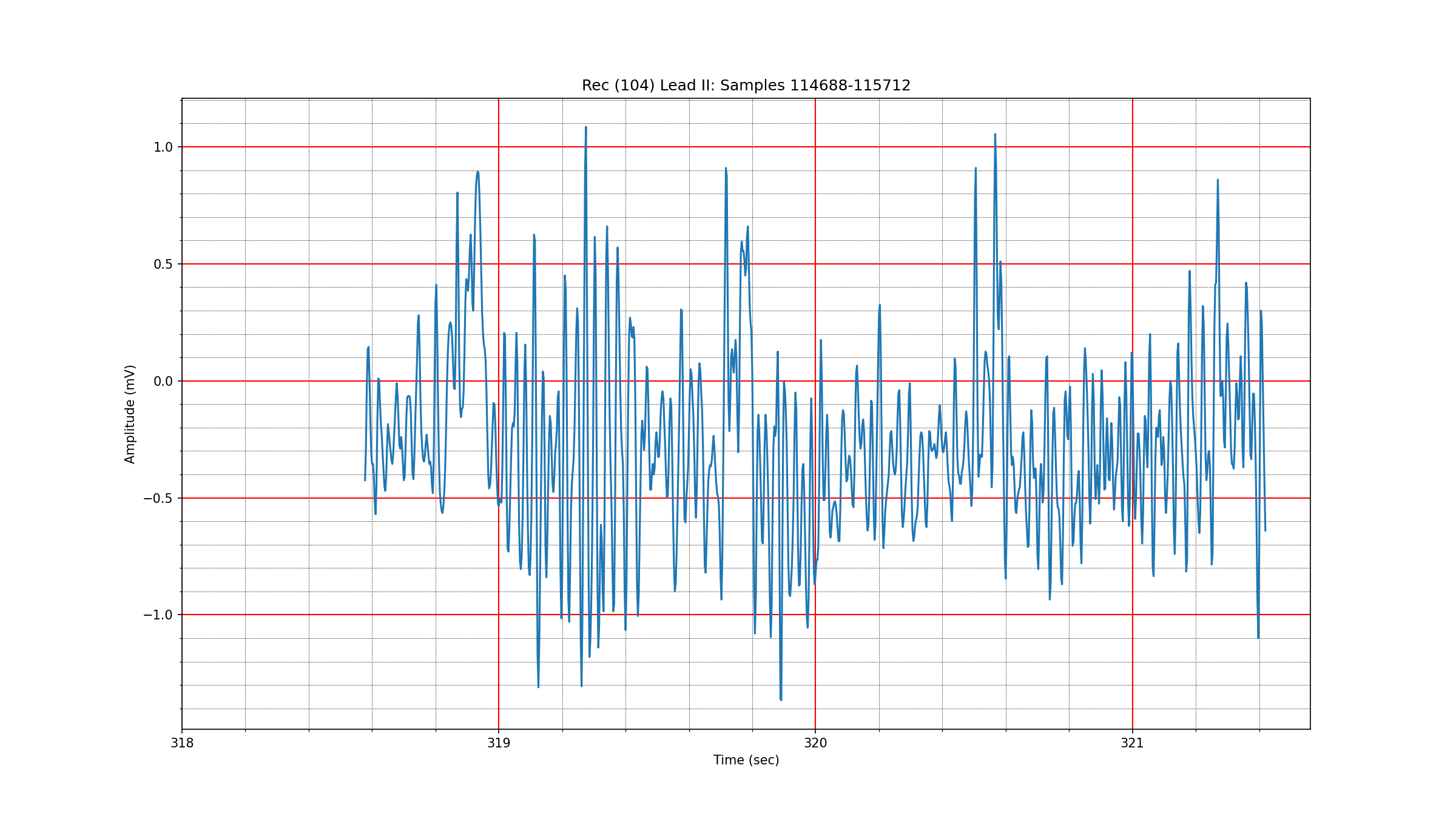
Fig. 3.7 An unclassified ECG segment from record 104#
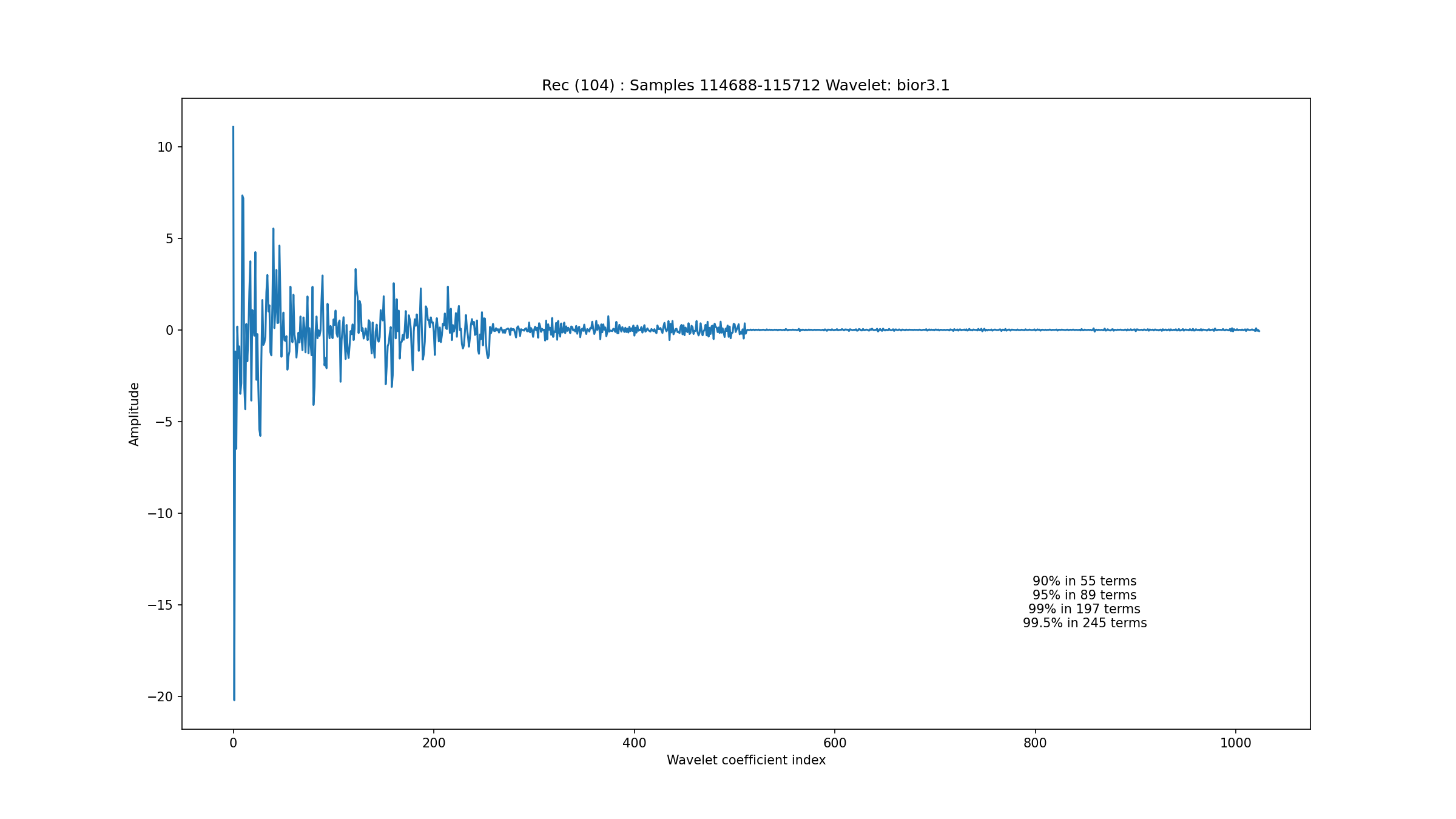
Fig. 3.8 The wavelet coefficients for the unclassified ECG segment from record 104#
3.4.8. Sparsity Variation for Different Beat Types#
We matched the beat annotations in the dataset with the corresponding 1024 sample blocks and obtained the corresponding sparsity level for 99% energy preservation. We examined the variation of sparsity level in different blocks for individual beat types. This data is provided in Table 3.6. It is interesting to see the significant amount of variation in the sparsity levels for pretty much all beat types. For some beat types, there are too few occurrences to look at the variation in sparsity levels.
sym |
counts |
|||||
|---|---|---|---|---|---|---|
0 |
N |
75052 |
7 |
159 |
36.8 |
11.4 |
1 |
L |
8075 |
9 |
127 |
33.3 |
8.8 |
2 |
R |
7259 |
12 |
134 |
37.0 |
11.9 |
3 |
V |
7130 |
9 |
137 |
33.8 |
9.0 |
4 |
/ |
7028 |
15 |
158 |
36.5 |
10.6 |
5 |
A |
2546 |
10 |
130 |
44.6 |
13.3 |
6 |
f |
982 |
17 |
197 |
41.0 |
17.2 |
7 |
F |
803 |
9 |
97 |
38.4 |
7.7 |
8 |
j |
229 |
18 |
152 |
39.9 |
17.8 |
9 |
a |
150 |
16 |
54 |
33.0 |
6.8 |
10 |
E |
106 |
15 |
47 |
23.2 |
5.5 |
11 |
J |
83 |
19 |
56 |
35.9 |
10.9 |
12 |
Q |
33 |
14 |
197 |
88.1 |
71.2 |
13 |
e |
16 |
24 |
37 |
31.5 |
4.2 |
14 |
S |
2 |
47 |
47 |
47.0 |
0.0 |