Architecture
Contents
4.1. Architecture#
4.1.1. Problem Statement#
We consider the problem of efficient transmission of
compressive measurements of ECG signals over the wireless body
area networks under the digital compressive sensing paradigm.
Let
We consider whether a digital quantization of the compressive measurements affects the reconstruction quality. Further, we study the empirical distribution of compressive measurements to explore efficient ways of entropy coding of the quantized measurements.
A primary constraint in our design is that the encoder should avoid any floating point arithmetic so that it can be implemented efficiently in low-power devices.
4.1.2. Block Diagram#
Fig. 4.1 presents the high-level block diagram of the proposed encoder.
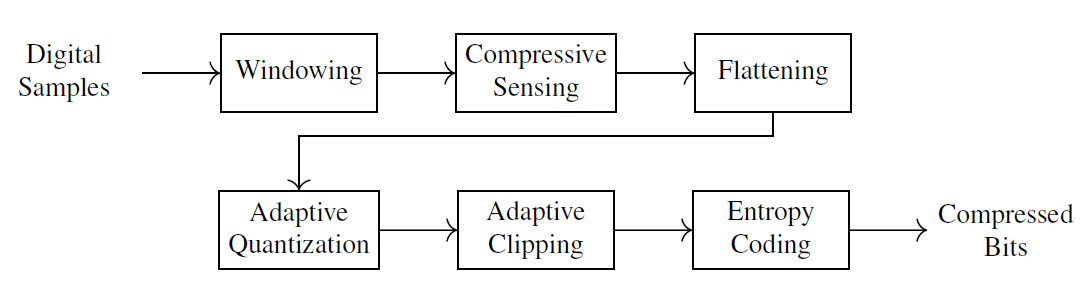
Fig. 4.1 Digital Compressive Sensing Encoder#
The ECG signal is split into windows
of
A window of the signal is the level at which the compressive sensing is done. A frame (a sequence of windows) is the level at which the adaptive quantization is applied and compressed signal statistics are estimated.
The encoding algorithm is further detailed in Algorithm 4.2.
Fig. 4.2 is a high-level block diagram of the proposed decoder.
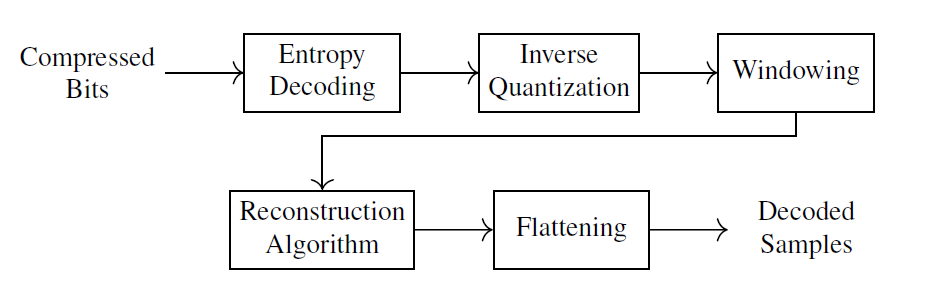
Fig. 4.2 Digital Compressive Sensing Decoder#
The decoding algorithm is further detailed in Algorithm 4.3.
4.1.3. Bitstream Format#
The encoder first sends the encoder parameters in the form of a stream header (Table 4.1). Then for each frame of the ECG signal, it sends a frame header (Table 4.2) followed by a frame payload consisting of the entropy-coded measurements for the frame.
Algorithm 4.1 (Bitstream format)
Send stream header.
While there are more ECG data frames:
Send frame header.
Send encoded frame data.
4.1.3.1. Stream Header#
The stream header Table 4.1
consists of all the necessary information
required to initialize the decoder.
In particular, it contains the pseudo-random
generator key that can be used to reproduce
the sparse binary sensing matrix used in the
encoder by the decoder,
the number of samples per window (
Parameter |
Description |
Bits |
|---|---|---|
key |
PRNG key for |
64 |
Window size |
12 |
|
Number of measurements per window |
12 |
|
Number of ones per column in sensing matrix |
6 |
|
Number of windows per frame |
8 |
|
adaptive |
Adaptive or fixed quantization flag |
1 |
if (adaptive ) |
||
|
8 |
|
else |
||
|
Fixed quantization parameter |
4 |
8 |
4.1.3.2. Frame Header#
The frame header precedes the encoded data for each frame. It captures the encoding parameters that vary from frame to frame.
Parameter |
Description |
Bits |
|---|---|---|
Mean value |
16 |
|
Standard deviation |
16 |
|
Frame quantization parameter |
3 |
|
Frame range parameter |
4 |
|
Windows in frame |
8 |
|
Words of entropy coded data |
16 |
4.1.4. Encoder#
Algorithm 4.2 (Encoder algorithm)
Send stream header.
Build sensing matrix
For each frame of ECG signal as
Compressive sensing:
Adaptive quantization. For
If
Quantized Gaussian model parameters
Adaptive range adjustment. For
If
Send frame header(
Send frame payload(
Here we describe the encoding process for each frame.
Fig. 4.1 shows a block diagram of the frame encoding process.
The input to the encoder is a frame of digital ECG samples
at a given sampling rate
4.1.4.1. Windowing#
A frame of ECG consists of multiple
such windows (up to a maximum of
PhysioNet provides the baseline values for each channel
in their ECG records.
Since the digital samples are unsigned, we have subtracted
them by the baseline value (
4.1.4.2. Compressive Sensing#
Following [22],
we construct a sparse binary sensing matrix
The digital compressive sensing operation is represented as
where
Note that by design, the sensing operation can be implemented
using just lookup and integer addition. The ones
in each row of
4.1.4.3. Flattening#
Beyond this point, the window structure of the signal is not
relevant for quantization and entropy coding purposes in our design.
Hence, we flatten it (column by column) into a vector
4.1.4.4. Quantization#
Next, we perform a simple quantization of measurement values.
If fixed quantization has been specified in the stream header,
then for each entry
For the whole measurement vector, we can write this as
This can be easily implemented in a computer as a signed
right shift by
If adaptive quantization has been specified, then we vary
the quantization parameter
4.1.4.5. Entropy Model#
Before we explain the clipping step, we shall describe
our entropy model.
We model the measurements as samples from a quantized Gaussian
distribution which can only take integral values.
First, we estimate the mean
Entropy coding works with a finite alphabet.
Accordingly, the quantized Gaussian model
requires the specification of the minimum
and maximum values that our quantized
measurements can take. The range of values
in
4.1.4.6. Adaptive Clipping#
The clipping function for scalar values is defined as follows:
We clip the values in
Similar to adaptive quantization, we vary
4.1.4.7. Entropy Coding#
We then model the measurement values in
4.1.4.8. Integer Arithmetic#
The input to digital compressive sensing is a stream of integer-valued ECG samples. The sensing process with the sparse binary sensing matrix can be implemented using integer sums and lookup. It is possible to implement the computation of approximate mean and standard deviation using integer arithmetic. We can use the normalized mean square error-based thresholds for adaptive quantization and clipping steps. ANS entropy coding is fully implemented using integer arithmetic. Thus, the proposed encoder can be fully implemented using integer arithmetic.
4.1.5. Decoder#
The decoder initializes itself by reading the stream header and creating the sensing matrix to be used for the decoding of compressive measurements frame by frame.
Algorithm 4.3 (Decoder algorithm)
Read stream header.
Build sensing matrix
While there is more data
Compute entropy model parameters
Inverse quantization:
Here we describe the decoding process for each frame.
Fig. 4.2 shows a block diagram for the
decoding process.
Decoding of a frame starts by reading the frame header
which provides the frame encoding parameters:
4.1.5.1. Entropy Decoding#
The frame header is used for building the quantized Gaussian distribution model for the decoding of the entropy-coded measurements from the frame payload. The minimum and maximum values for the model are computed as:
4.1.5.2. Inverse Quantization#
We then perform the inverse quantization as
Next, we split the measurements into measurement windows of size
We are now ready for the reconstruction of the ECG signal for each window.
4.1.5.3. Reconstruction#
The architecture is flexible in terms of the choice of the reconstruction algorithm.
Each column (window) in
4.1.5.4. Alternate Reconstruction Algorithms#
It is entirely possible to use a deep learning-based
reconstruction network like CS-NET [32]
in the decoder. We will need to train the network with
4.1.6. Discussion#
4.1.6.1. Measurement statistics#
Several aspects of our encoder architecture are based on the
statistics of the measurements
rec |
iqr |
rng |
skew |
kurtosis |
kld |
||||
|---|---|---|---|---|---|---|---|---|---|
100 |
-490 |
224 |
293 |
2562 |
-0.46 |
3.71 |
0.05 |
11.41 |
1.31 |
101 |
-455 |
287 |
323 |
9647 |
-0.39 |
15.4 |
0.13 |
33.61 |
1.13 |
102 |
-393 |
197 |
257 |
2035 |
-0.55 |
3.68 |
0.05 |
10.33 |
1.31 |
103 |
-370 |
286 |
302 |
8824 |
-0.78 |
15.91 |
0.16 |
30.86 |
1.06 |
104 |
-360 |
231 |
284 |
4204 |
-0.61 |
5.04 |
0.07 |
18.16 |
1.23 |
105 |
-360 |
347 |
326 |
13704 |
-0.38 |
22.52 |
0.24 |
39.52 |
0.94 |
106 |
-285 |
271 |
314 |
4135 |
-0.09 |
4.43 |
0.05 |
15.28 |
1.16 |
107 |
-372 |
625 |
747 |
9639 |
-0.46 |
4.65 |
0.06 |
15.43 |
1.2 |
108 |
-366 |
422 |
336 |
12986 |
-0.16 |
22.13 |
0.36 |
30.75 |
0.8 |
109 |
-368 |
335 |
389 |
7487 |
0.19 |
7.33 |
0.06 |
22.32 |
1.16 |
111 |
-262 |
308 |
306 |
11998 |
-0.91 |
23.07 |
0.2 |
38.99 |
0.99 |
112 |
-1316 |
539 |
724 |
7839 |
-0.64 |
4.14 |
0.09 |
14.55 |
1.34 |
113 |
-248 |
337 |
379 |
6121 |
-0.05 |
5.19 |
0.06 |
18.18 |
1.13 |
114 |
-249 |
219 |
235 |
8756 |
1.15 |
30.41 |
0.14 |
39.91 |
1.07 |
115 |
-781 |
461 |
559 |
9715 |
-0.82 |
6.74 |
0.09 |
21.07 |
1.21 |
116 |
-1498 |
875 |
930 |
35083 |
-0.29 |
22.7 |
0.22 |
40.1 |
1.06 |
117 |
-1363 |
560 |
748 |
9179 |
-0.7 |
4.38 |
0.1 |
16.39 |
1.34 |
118 |
-1373 |
614 |
809 |
10935 |
-0.55 |
4.37 |
0.1 |
17.81 |
1.32 |
119 |
-1378 |
629 |
825 |
8363 |
-0.57 |
3.8 |
0.07 |
13.3 |
1.31 |
121 |
-1296 |
635 |
763 |
17012 |
-1.37 |
13.12 |
0.16 |
26.78 |
1.2 |
122 |
-1350 |
555 |
747 |
7870 |
-0.55 |
3.73 |
0.08 |
14.18 |
1.35 |
123 |
-1274 |
514 |
699 |
6603 |
-0.57 |
3.83 |
0.08 |
12.84 |
1.36 |
124 |
-1293 |
679 |
829 |
11886 |
-0.6 |
5.42 |
0.1 |
17.51 |
1.22 |
200 |
-169 |
262 |
288 |
6285 |
-0.26 |
7.71 |
0.08 |
24.01 |
1.1 |
201 |
-256 |
189 |
196 |
9986 |
0.72 |
55.1 |
0.16 |
52.71 |
1.03 |
202 |
-271 |
262 |
274 |
11421 |
-0.98 |
32.98 |
0.16 |
43.58 |
1.05 |
203 |
-271 |
472 |
455 |
13371 |
-0.14 |
15.11 |
0.2 |
28.35 |
0.96 |
205 |
-489 |
234 |
301 |
3257 |
-0.57 |
4.1 |
0.06 |
13.94 |
1.29 |
207 |
-274 |
339 |
344 |
11400 |
-1.19 |
18.15 |
0.22 |
33.62 |
1.01 |
208 |
-265 |
427 |
410 |
12449 |
0.38 |
16.51 |
0.18 |
29.13 |
0.96 |
209 |
-264 |
250 |
292 |
4731 |
-0.55 |
4.89 |
0.06 |
18.95 |
1.17 |
210 |
-251 |
228 |
249 |
7909 |
0.48 |
17.22 |
0.09 |
34.65 |
1.09 |
212 |
-250 |
297 |
336 |
7232 |
-0.53 |
7.26 |
0.09 |
24.36 |
1.13 |
213 |
-353 |
523 |
635 |
6528 |
-0.18 |
4.03 |
0.04 |
12.48 |
1.21 |
214 |
-258 |
315 |
369 |
4248 |
0.16 |
4.12 |
0.04 |
13.47 |
1.17 |
215 |
-243 |
209 |
256 |
3544 |
-0.27 |
4.2 |
0.04 |
16.98 |
1.23 |
217 |
-264 |
459 |
528 |
11928 |
-0.3 |
8.5 |
0.09 |
26 |
1.15 |
219 |
-939 |
688 |
811 |
10727 |
-0.43 |
4.63 |
0.07 |
15.6 |
1.18 |
220 |
-865 |
378 |
503 |
4731 |
-0.48 |
3.66 |
0.06 |
12.5 |
1.33 |
221 |
-262 |
228 |
270 |
3000 |
-0.13 |
4.1 |
0.04 |
13.16 |
1.18 |
222 |
-250 |
212 |
252 |
4327 |
-0.78 |
5.71 |
0.08 |
20.41 |
1.19 |
223 |
-818 |
419 |
529 |
7449 |
-0.58 |
4.95 |
0.07 |
17.77 |
1.26 |
228 |
-222 |
357 |
294 |
11962 |
-0.97 |
23.96 |
0.35 |
33.52 |
0.82 |
230 |
-272 |
285 |
308 |
7044 |
-0.4 |
8.63 |
0.1 |
24.69 |
1.08 |
231 |
-253 |
229 |
278 |
3250 |
-0.34 |
4.21 |
0.04 |
14.2 |
1.21 |
232 |
-242 |
192 |
243 |
2951 |
-0.42 |
3.98 |
0.04 |
15.38 |
1.27 |
233 |
-246 |
427 |
480 |
9636 |
-0.13 |
6.47 |
0.08 |
22.56 |
1.12 |
234 |
-257 |
345 |
316 |
12002 |
-0.66 |
19.78 |
0.25 |
34.78 |
0.92 |
4.1.6.2. Gaussianity#
The key idea behind our entropy coding
design is to model the measurement values as
being sampled from a quantized
Gaussian distribution. Towards this, we measured the
skew and kurtosis for the measurements for each record
as shown in Table 4.3
for the sensing matrix configuration of
The best compression can be achieved by using the empirical
probabilities of different values in
We computed the empirical distribution for
Fig. 4.9 shows an example
where the empirical distribution is significantly different
from the corresponding quantized Gaussian distribution
due to the presence of a large
number of
Also, Fig. 4.3-Fig. 4.8 suggest that the empirical distributions vary widely from one record to another in the database. Hence using a single fixed empirical distribution (e.g. the Huffman code-book preloaded into the device in [22]) may lead to lower compression.
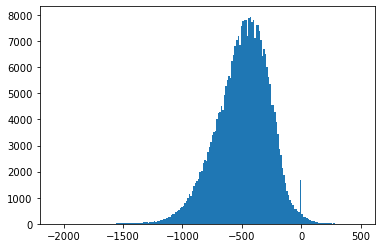
Fig. 4.3 Histograms of measurement values for record 100#
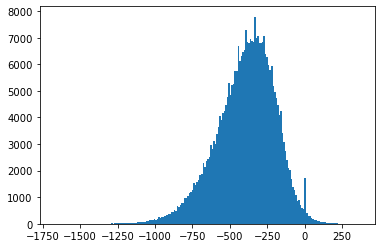
Fig. 4.4 Histograms of measurement values for record 102#
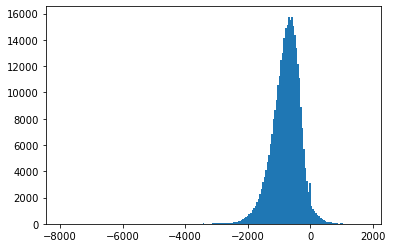
Fig. 4.5 Histograms of measurement values for record 115#
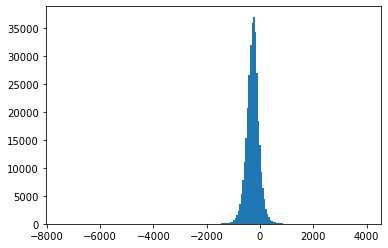
Fig. 4.6 Histograms of measurement values for record 202#
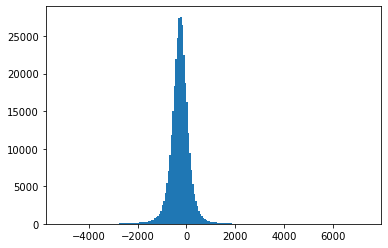
Fig. 4.7 Histograms of measurement values for record 208#
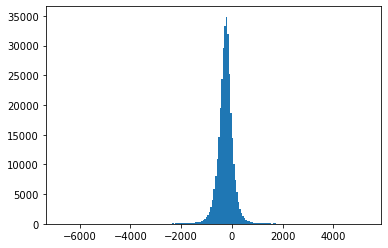
Fig. 4.8 Histograms of measurement values for record 234#
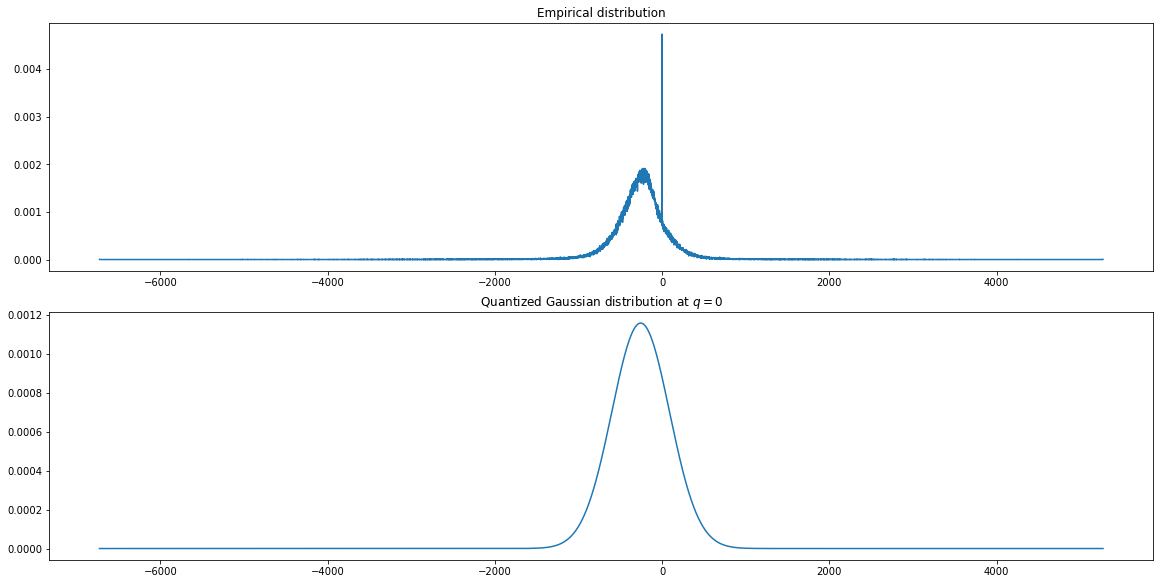
Fig. 4.9 Empirical and quantized Gaussian distributions for measurement values
4.1.6.3. Clipping#
An entropy coder can handle a finite set of symbols
only. Hence, the range of input values [measurements
coded as integers] must be restricted to a finite range.
This is the reason one has to choose a distribution
with finite support (like quantized Gaussian).
From Table 4.3 one can see that
while the complete range of measurement values can be
up to 40-50x larger than the standard deviation, the iqr
is less than
4.1.6.4. Quantization#
Fig. 4.10 demonstrates the impact of the
quantization step on the reconstruction quality
for record 100 under non-adaptive quantization.
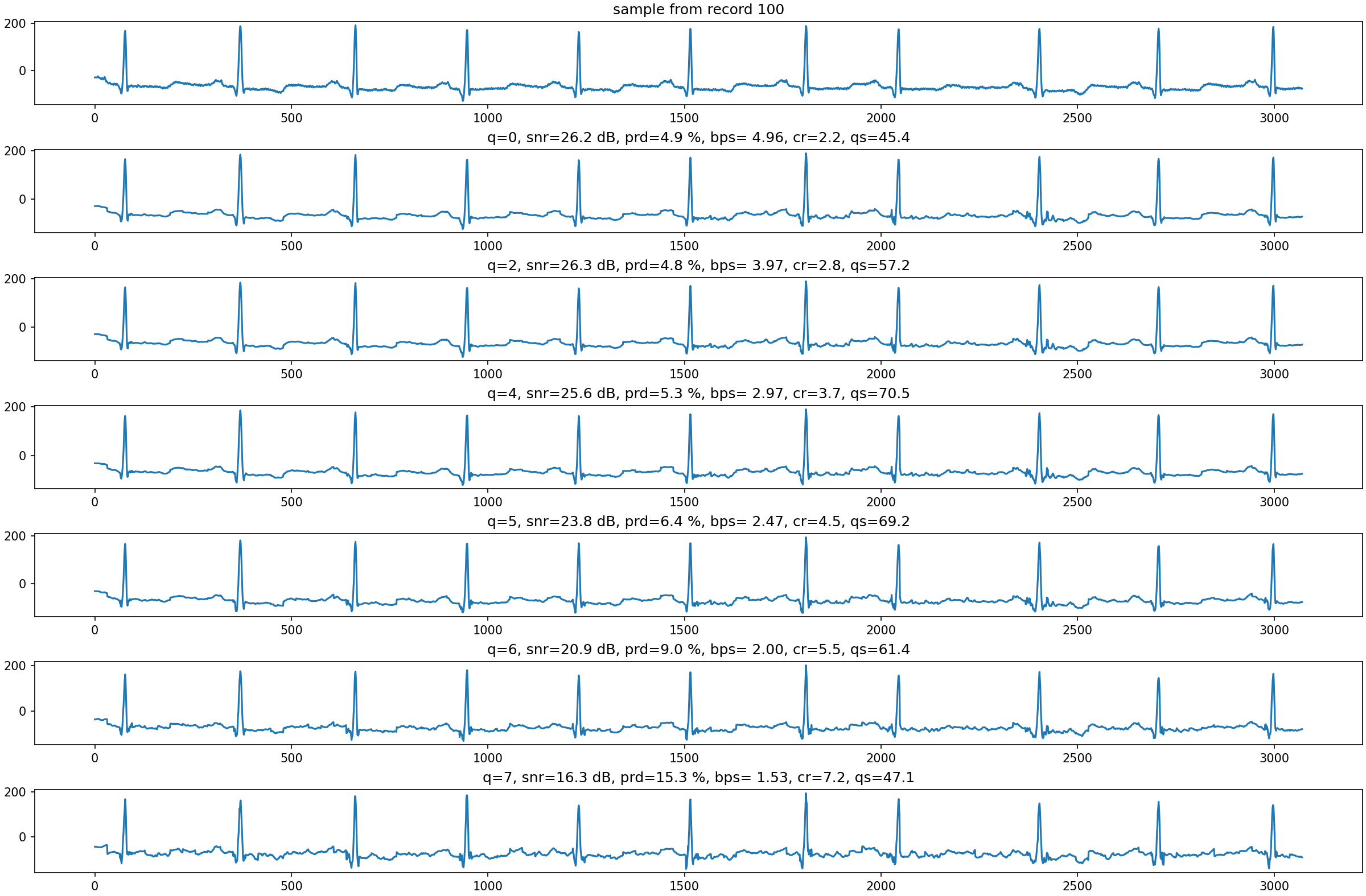
Fig. 4.10 Reconstruction of a small segment of record 100
for different values of